Overview
Major societal and environmental challenges require forecasting how natural processes and human activities affect one another. Model integration across natural and social science disciplines to study these problems requires resolving semantic, spatio-temporal, and execution mismatches, which are largely done by hand today and may take more than two years of human effort.
We are developing the Model INTegration (MINT) framework that incorporates extensive knowledge about models and data, with several innovative components:
- New principle-based ontology generation tools for modeling variables, used to describe models and data;
- A novel workflow system that selects relevant models from a curated registry and uses abductive reasoning to hypothesize new models and data transformation steps;
- A new data discovery and integration framework that finds and categorizes new sources of data, learns to extract information from both online sources and remote sensing data, and transforms the data into the format required by the models;
- New knowledge-guided machine learning algorithms for model parameterization to improve accuracy and estimate uncertainty;
- A novel framework for multi-modal scalable workflow execution.
We are building on many previously existing tools, including CSDMS, BMI, GSN, WINGS, Pegasus, Karma, and GOPHER. Rapid model integration would enable efficient and comprehensive coupled human and natural system modeling.
Ontologies
A major challenge in integrating models is understanding model variables, processes, and assumptions. For example, a model may refer to "streamflow" and another to “discharge” and it may take some time to understand that they refer to the same physical variable. Although standards and ontologies have been created for specific domains, mapping variables across them remains an open problem. Our approach is to develop general principles and turn them into patterns to create names for model variables, processes, and assumptions. In prior work, we developed a cross-domain ontology called the Geoscience Standard Names (GSN) [1]. The GSN ontology was designed to serve as a semantic mediation hub and is based on very general principles that have been shown to apply to a wide variety of science domains including oceanography, atmospheric science, hydrology, glaciology, sea ice, geomorphology, general physics, continuum mechanics, thermodynamics, electricity and magnetism, seismology and environmental chemistry. GSN also includes standards for assumptions that models make, such as the Navier-Stokes equation for fluid dynamics.
Leads
Dr. Peckham is a Senior Research Scientist at INSTAAR at the University of Colorado, Boulder. His research is in hydrology and fluvial landscape evolution, with expertise in fluid dynamics, digital terrain analysis, mathematical modeling, scaling theory, stochastic processes, software development and cyber-infrastructure.
References
- Peckham, S.D. 2014. The CSDMS Standard Names: Cross-domain naming conventions for describing process models, data sets and their associated variables. Proceedings of the 7th Intl. Congress on Env. Modelling and Software, International Environmental Modelling and Software Society (iEMSs).
Modeling
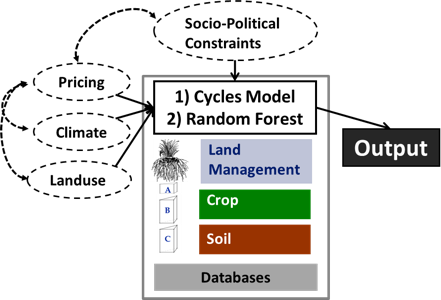
The MINT project team will address primarily food production under changing scenarios, changes in the water cycle due to external forcing such as climate or market changes, and internal changes such as changes in food and water demand due to land use change including urban growth and purchasing power. Food production will be predicted based on process-based models (model Cycles) that are able to use daily or sub-daily climate information, and machine learning (Random Forest). The Cycles model is an evolution of C-Farm [1] and shares many modules with CropSyst [2]. The Cycles model simulates yield and environmental impacts of food production based on fundamental biophysical principles that control plant growth, water and nutrient cycling. Its modular structure, transparent input and output structure, documentation in Github, and power to model any crop and crop rotation make the Cycles model an ideal component for integration in a modeling workflow alongside other models. Unlike process based models, machine learning algorithms can predict single indicators faster and using multiple sources of information, regardless of the specific mechanistic connection between predictor and predicted variable. They can be particularly powerful to predict crop production [3].
A challenge to integrating economic with biophysical models is a lack of standardized, portable modeling platforms for the former. For this project, I am developing modular and reusable economic models of land-use activities and land-use change that can be used to facilitate efficient bio-economic modeling. Building on microeconomic theory and established quantitative modeling approaches, I develop modules that represent landowner decision-making in cropping activities, livestock production, native forest harvesting, and plantation forest management. These modules are nested within a predictive model of land-use change based on a comparison of the relative economic rents to land in each activity. The core models are extensible to include property rights insecurity that arises from expropriation risk and illegal logging, as well as the effects of migration on land rents.
Leads
Dr. Kemanian has background in agroecology, systems modeling, and several disciplines central to agricultural and natural systems. He developed the Cycles model, components of the CropSyst model, and made contributions to the SWAT model and associated models EPIC and APEX. He is currently using the PIHM and Hydroterre platform, migrating to fully distributed models to represent terrestrial and aquatic processes. These models have been used in numerous projects of local, national and international reach.
Dr. Duffy is a Professor in the Civil and Environmental Engineering Department Penn State University in the area of Water Resources Engineering. Currently Dr. Duffy and his team have focused on developing the spatially-distributed, physics-based computational model PIHM (The Penn State Integrated Hydrologic Model) for multi-scale, multi-process water resources applications, and an on-line national data service for access to geospatial watershed data anywhere in the continental US.
Dr. Cobourn is an Assistant Professor of Water Economics and Policy in the Department of Forest Resources and Environmental Conservation at Virginia Tech. She conducts research into integrated hydroeconomic and bioeconomic modeling using dynamic programming techniques and the use of data-driven econometric methods and positive mathematical programming (PMP) to parameterize and calibrate integrated models.
References
- Kemanian, A.R. and Stöckle, C.O., 2010. C-Farm: A simple model to evaluate the carbon balance of soil profiles. European Journal of Agronomy, 32(1), pp.22-29.
- Stöckle, C.O., Kemanian, A.R., Nelson, R.L., Adam, J.C., Sommer, R. and Carlson, B., 2014. CropSyst model evolution: From field to regional to global scales and from research to decision support systems. Environmental Modelling & Software, 62, pp.361-369.
- Hoffman, A.L., Kemanian, A.R. and Forest, C.E., 2018. Analysis of climate signals in the crop yield record of sub‐Saharan Africa. Global Change Biology, 24(1), pp.143-157.
Model Calibration
Since the models being considered in this project vary in their degree of completeness, uncertainty of knowledge, and degree of validation, many models use parametric forms of approximations for processes that are not well-understood and suffer from large knowledge gaps. In such settings, domain-specific models are often forced to make a number of inexact approximations about the physical processes, which not only leads to poor predictive performance but also renders the model difficult to comprehend and analyze. To address these issues, we will build upon the paradigm of theory-guided data science (TGDS) that we have recently formulated [1, 2, 3]. This approach introduces scientific consistency as an essential component for learning generalizable models. We will use TGDS to develop novel methods for model parameterization, where both physics and data science are used in a synergistic manner in hybrid-physics-data models.
Lead
Dr. Kumar is a Regents Professor and holds William Norris Chair in the department of Computer Science and Engineering at the University of Minnesota. Dr. Kumar's current research interests include data mining, high-performance computing, and their applications in Climate/Ecosystems and health care. His research has resulted in the development of the concept of isoefficiency metric for evaluating the scalability of parallel algorithms, as well as highly efficient parallel algorithms and software for sparse matrix factorization (PSPASES) and graph partitioning (METIS, ParMetis, hMetis).
References
- A. Karpatne, G. Atluri, J. Faghmous, M. Steinbach, A. Banerjee, A. Ganguly, S. Shekhar, N. Samatova, and V. Kumar, 2017. Theory-guided data science: A new paradigm for scientific discovery. In IEEE Transactions on Knowledge and Data Engineering, 29(10).
- J. H. Faghmous, A. Banerjee, S. Shekhar, M. Steinbach, V. Kumar, A. R. Ganguly, and N. Samatova, 2014. Theory-guided data science for climate change. Computer, 47(11):74–78.
- J. H. Faghmous and V. Kumar, 2014. A Big Data Guide to Understanding Climate Change: The Case for Theory-Guided Data Science. Big Data, 2(3).
Data Extraction and Integration
Leads
Dr. Knoblock is a Research Professor of both Computer Science and Spatial Sciences at the University of Southern California (USC), Research Director of Information Integration at the Information Sciences Institute, and Associate Director of the Informatics Program at USC. His research focuses on techniques for describing, acquiring, and exploiting the semantics of data. He has worked extensively on source modeling, schema and ontology alignment, entity and record linkage, data cleaning and normalization, extracting data from the Web, and combining these techniques to build knowledge graphs. He has also worked specifically on the challenges of aggregating and integrating country-level statistical data related to food security from government agencies (including the United Nations, World Health Organization) and NGOs.
Workflow Composition
To generate workflows composed of diverse models and the necessary data transformation steps, we use the WINGS semantic workflow system [1] to reason about data characteristics and model requirements available in the MINT Model and Data Catalogs. While a traditional workflow simply represents dataflow among software components, a semantic workflow also represents the characteristics of the input and output datasets for each software step and any constraints in those datasets or parameters to the step. WINGS includes workflow reasoning algorithms that propagate those constraints for automated workflow elaboration, workflow matching, provenance and metadata generation, workflow validation, and interactive assistance. A user would interact with the MINT throughout the workflow planning process. The user starts by specifying some variables of interest, which would indicate the scope of the problem and the level of detail required of the models. Those variables are then mapped to the MINT ontologies, and used to select relevant models. Each model grouping would represent the initial skeleton for a workflow, which would then be expanded using the data pre-processing workflow fragments specified in the Model Catalog. This results in an initial workflow template. WINGS will then reason about the requirements of each model and add any data conversion steps needed to transform model outputs into the format required by other models.
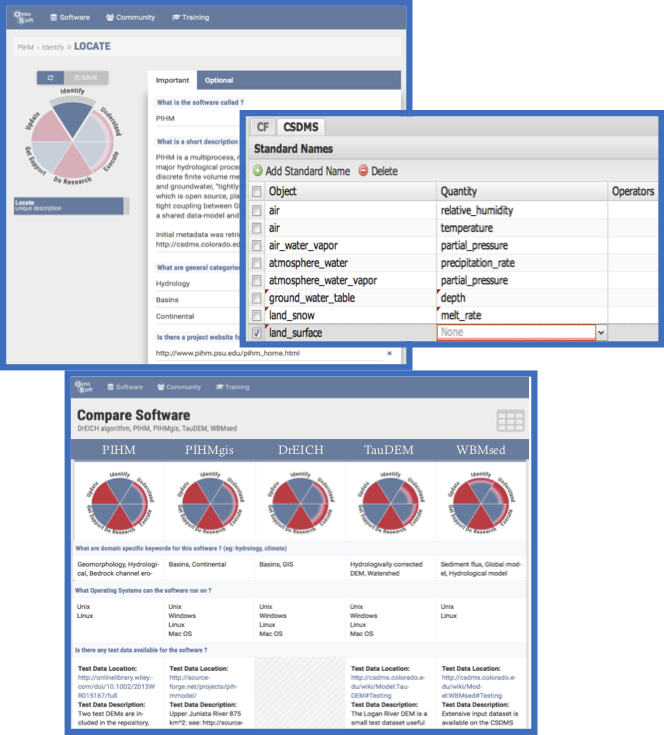
MINT is developing a model repository with machine-actionable model metadata that can be used to provide intelligent assistance to scientists in model selection and reuse. The MINT model catalog extends the OntoSoft semantic software metadata registry to include machine-readable metadata. This work includes: 1) exposing model variables and their relationships; 2) exposing model processes and how they group and relate to model variables; 3) adopting a standardized representation of model variables based on the conventions of the Geoscience Standard Names ontology (GSN); 4) capturing the semantic structure of model invocation signatures based on functional inputs and outputs and their correspondence to model variables; 5) associating models with readily reusable workflow fragments for data preparation, model calibration, and visualization of results.
Lead
Dr. Gil is a Director of Knowledge Technologies at USC's Information Sciences Institute (ISI), and a Research Professor in Computer Science and Spatial Sciences. Her research is on intelligent interfaces for knowledge capture and discovery, which she investigates in a variety of projects concerning knowledge-based planning and problem solving, information analysis and assessment of trust, semantic annotation and metadata, and community-wide development of knowledge bases. Dr. Gil collaborates with scientists in different domains on semantic workflows and metadata capture, social knowledge collection, computer-mediated collaboration, and automated discovery.
References
- Mind Your Metadata: Exploiting Semantics for Configuration, Adaptation, and Provenance in Scientific Workflows. Gil, Y.; Szekely, P.; Villamizar, S.; Harmon, T.; Ratnakar, V.; Gupta, S.; Muslea, M.; Silva, F.; and Knoblock, C. In Proceedings of the Tenth International Semantic Web Conference (ISWC), Bonn, Germany 2011.
- Gil, Y., Ratnakar, V., Kim, J., Gonzalez-Calero, P., Groth, P., Moody, J. and Deelman, E., 2011. Wings: Intelligent workflow-based design of computational experiments. IEEE Intelligent Systems, 26(1), pp.62-72.
Workflow Execution
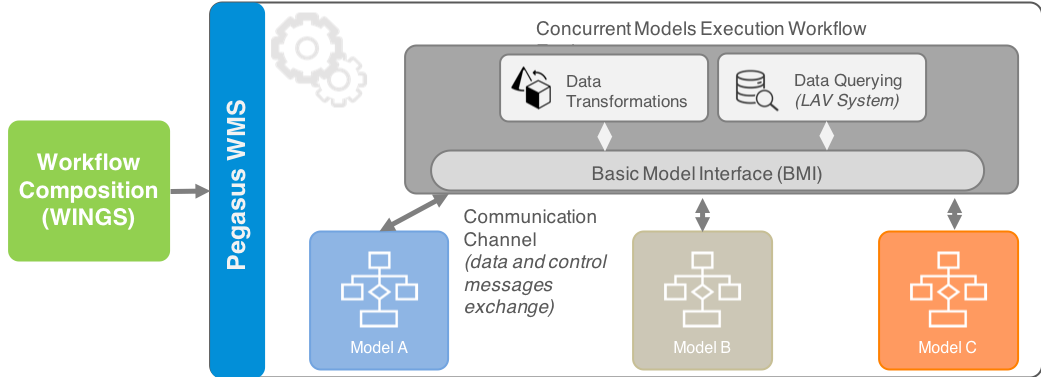
Workflows process scientific large-scale computations in distributed systems. A key advantage of workflows is their ability to combine different specialized software. Typically, they are described as directed-acyclic graphs (DAGs), where the outputs of a job (a node in the graph) are input for subsequent jobs. This paradigm fits model chaining workflows, where each model runs once and its output is input for successor models. The MINT team will capitalize on our Pegasus [1] workflow system to enable scalable workflow execution in computing environments. Although DAGs are proven efficient for most scientific computations, they are insufficient to handle coupled models – concurrent execution of models with continuous data exchanges. We will leverage our prior work on the CSDMS Basic Model Interface (BMI) that provides standardized, noninvasive, and framework-independent API for models [3]. BMI is easy to implement and yet provides all information needed to deploy a model in multiple model coupling frameworks.
Leads
Dr. Deelman is a Research Professor at the USC Computer Science Department and a Research Director at the USC Information Sciences Institute. Dr. Deelman’s research interests include the design and exploration of collaborative, distributed scientific environments, with particular emphasis on workflow management as well as the management of large amounts of data and metadata. At ISI, Dr. Deelman is leading the Pegasus project, which designs and implements workflow mapping techniques for large-scale applications running in distributed environments. Pegasus is being used today in a number of scientific disciplines, enabling researches to formulate complex computations in a declarative way. Dr. Deelman received her Ph.D. in Computer Science from the Rensselaer Polytechnic Institute in 1997. Her thesis topic was in the area of parallel discrete event simulation, where she applied parallel programming techniques to the simulation of the spread of Lyme disease in nature.
Dr. Ferreira da Silva is a Research Assistant Professor in the Department of Computer Science at University of Southern California, and a Computer Scientist in the Science Automation Technologies group at the USC Information Sciences Institute. His research focuses on the efficient execution of scientific workflows on heterogeneous distributed systems (e.g., clouds, grids, and supercomputers), computational reproducibility, and Data Science workflow performance analysis, user behavior in HPC/HTC, and citation analysis (for publications). Dr. Ferreira da Silva received his Ph.D. in Computer Science from INSA Lyon, France, in 2013. He authored more than 60 research papers in the are of distributed computing, in particular high performance computing and workflows.
References
- Deelman, E., et al. "Pegasus, a workflow management system for science automation." Future Generation Computer Systems 46 (2015): 17-35.
- Peckham, Scott D., Eric WH Hutton, and Boyana Norris. "A component-based approach to integrated modeling in the geosciences: The design of CSDMS." Computers & Geosciences 53 (2013): 3-12.
